Abstract
Introduction: Systems biology approaches can identify critical dependencies in complex cancer signaling networks to inform rational therapy combinations and overcome treatment resistance (Hu Nat. Comm. 2019). Philadelphia chromosome-like B-acute lymphoblastic leukemia (Ph-like ALL) is a common high-risk leukemia subtype defined by a kinase-activated gene expression pattern and chemoresistance with low overall survival rates in both children and adults. Clinical responses of patients with Ph-like ALL to tyrosine kinase inhibitor (TKI)-based therapies may be incomplete, and alternative biologic dependencies remain incompletely elucidated. We previously reported the therapeutic potential of combined JAK and BCL-2 inhibition in the most common CRLF2-rearranged (R) subtype of Ph-like ALL (Ding CCR 2021). Herein, we report integrated bulk and single-cell (sc) multi-omics studies of Ph-like ALL cells treated in vitro and in vivo with ruxolitinib to define transcriptional rewiring that leads to TKI escape.
Methods: To interrogate oncogenic dependencies and mechanisms of TKI evasion in CRLF2-R Ph-like ALL, we treated patient-derived xenograft (PDX) mouse models (n=4; 5 mice per treatment arm per model) with the JAK1/2 inhibitor (i) ruxolitinib or vehicle for up to 2 months as previously described (Tasian Blood 2017a). We performed and integrated bulk RNA-seq and ATAC-seq analyses of human ALL cells harvested from murine spleens to elucidate altered intracellular pathways and transcriptional regulatory network changes. To investigate if heterogeneity of drug-induced transcriptional states may contribute to drug resistance, we performed scRNA-seq and scATAC-seq of persisting leukemia cells after treatment in vivo with ruxolitinib and the BCL-2 inhibitor venetoclax.
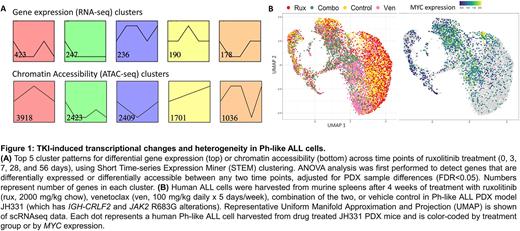
Results: We observed that long-term ruxolitinib treatment of Ph-like ALL PDX models resulted in a transcriptomic state shift in CRLF2-R ALL cells with thousands of differentially expressed genes and altered chromatin accessibility (Fig. 1A), suggesting global gene regulatory effects. Using time course RNA-seq and ATAC-seq data gathered from PDX ALL cells at early (3 and 7 days) and later (28 and 56 days) timepoints during ruxolitinib therapy, we identified pathways associated with potential TKI evasion, including cytokine signaling, cell cycling, cell death, and histone modification (Fig. 1A). We identified key transcription factors that mediate gene regulatory network rewiring associated with therapeutic escape, including MYC as a highly ranked mediator. We found potent inhibition of Ph-like ALL viability in vitro when combining a new MYC inhibitor MYCi975 (Han Cancer Cell 2019) with ruxolitinib at sub-IC50 doses. Given the above results, we postulated that MYC-mediated effects on cell cycling and dormancy may allow for sub-populations of 'persister cells' to develop TKI-resistant phenotypes. We integrated scRNA-seq and scATAC-seq data from residual human ALL cells harvested from PDX models after targeted inhibition with ruxolitinib, venetoclax (to target dormant cells), or both inhibitors in combination (representative data shown in Fig. 1B). We observed striking heterogeneity of MYC expression at a single-cell level and significant changes in functionally relevant transcriptional signatures for senescence and apoptosis. We observed an increased proportion of cells arrested in G0/G1 (with corresponding higher senescence-like score) during ruxolitinib monotherapy with more cells arrested in S phase (corresponding to a lower senescence-like score) during combined JAK and BCL-2 inhibition.
Conclusions: Our findings suggest that Ph-like ALL cells can resist ruxolitinib treatment via TKI-induced gene regulatory network rewiring that leads to altered apoptosis and dormant/senescent states. Ongoing studies are investigating the therapeutic potential of targeting these dormant cells using inhibitors such as venetoclax, senolytics, or novel MYC inhibitors, which may have translational potential for future combinatorial treatment of patients with Ph-like ALL.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal